I want to find indices of all matches of a vector x in another lookup vector table.
table = rep(1:5, each=3)
x = c(2, 5, 2, 6)
Standard base R methods don’t quite give me what I want. For example using which(table %in% x) we only get the matching indices once, even though 2 appears twice in x
which(table %in% x)
# [1] 4 5 6 13 14 15
On the other hand, match returns values for every occurrence of x that has a match, but only returns the first index in the lookup table.
match(x, table)
# [1] 4 13 4 NA
What I want is a function that returns the indices for "all x and all y". I.e. it should return the following desired result:
mymatch(x, table)
# c(4, 5, 6, 13, 14, 15, 4, 5, 6)
We can, of course, do this with a loop in R:
mymatch = function(x, table) {
matches = sapply(x, (xx) which(table %in% xx))
unlist(matches)
}
mymatch(x, table)
# [1] 4 5 6 13 14 15 4 5 6
But this is horribly slow on larger data (I need to do this operation many times on big data)
table = rep(1:1e5, each=10)
x = sample(1:100, 1000, replace = TRUE)
system.time(mymatch(x, table))
# user system elapsed
# 3.279 2.881 6.157
This is very slow if we compare it to e.g. which %in%:
system.time(which(table %in% x))
# user system elapsed
# 0.003 0.004 0.008
Hoping there is a fast way to do this in R? Otherwise, maybe RCpp is the way to go.
8
6 Answers
Another way is to use split:
unlist(split(seq(table), table)[as.character(x)],use.names = FALSE)
[1] 4 5 6 13 14 15 4 5 6
Edit:
Note that if table is sorted, then you could use rle + sequence:
faster <- function(x, table){
a <- rle(table)
n <- length(a$lengths)
idx <- match(x, a$values, 0)
sequence(a$lengths[idx], cumsum(c(1,a$lengths[-n]))[idx])
}
set.seed(42)
table = rep(1:1e5, each=10)
x = sample(1:100, 1000, replace = TRUE)
bench::mark(
faster(x, table),
#mymatch(x, table) |> as.vector(),
join_match(x, table),
#unlist(split(seq(table), table)[as.character(x)],use.names = FALSE),
check = TRUE
)
# A tibble: 2 × 13
expression min median `itr/sec` mem_alloc `gc/sec` n_itr n_gc total_time result memory
<bch:expr> <bch:t> <bch:> <dbl> <bch:byt> <dbl> <int> <dbl> <bch:tm> <list> <list>
1 faster(x,… 54.4ms 252ms 3.97 54.9MB 1.99 2 1 503ms <int> <Rprofmem>
2 join_matc… 127.7ms 254ms 3.93 88.8MB 5.90 2 3 508ms <int> <Rprofmem>
# ℹ 2 more variables: time <list>, gc <list>
The function works as long as table is sorted. Not necessarily from 1:n.
table = c(rep(1:5, each=3), 7,7,7,7,10,10)
x = c(10, 2, 5,7, 2, 6)
microbenchmark::microbenchmark(
faster(x, table),
#mymatch(x, table) |> as.vector(),
join_match(x, table),
#unlist(split(seq(table), table)[as.character(x)],use.names = FALSE),
check = 'equal'
)
Unit: microseconds
expr min lq mean median uq max neval
faster(x, table) 23.001 32.751 56.95703 56.400 66.201 222.901 100
join_match(x, table) 4216.201 4925.302 6616.51401 5572.951 7842.200 21153.402 100
4
-
1
I would like to award this the "most elegant solution", which is also impressively fast. But will probably give the check mark to the join version, because it is (slightly) faster. Worth noting though that the memory allocation for this is one third that of the
join_matchanswer, so if memory rather than speed were my bottleneck, I would use this solution.– dww23 hours ago
-
For my current use case, yes it is a sorted vector with n repetitions. But the values are not contiguous sequences. A faster solution that uses these constraints would certainly be great.
– dww23 hours ago
-
1
@dww check the edit
– Onyambu22 hours ago
-
2
@dww Note that if
xhas alot of repetitions within it, you rather only match the unique values and the extract for the specific x values. ie instead of doingmatch(c(2,2,2,2,2), table)justmatch(2,table)then index accordingly– Onyambu22 hours ago
Maybe data.table would be an option? If you have relatively large table/vector you might see an improvement in speed, especially if you go with something along the lines of Jon Spring’s "join" approach:
library(tidyverse)
library(data.table)
#>
#> Attaching package: 'data.table'
#> The following objects are masked from 'package:lubridate':
#>
#> hour, isoweek, mday, minute, month, quarter, second, wday, week,
#> yday, year
#> The following objects are masked from 'package:dplyr':
#>
#> between, first, last
#> The following object is masked from 'package:purrr':
#>
#> transpose
library(microbenchmark)
onyambu_faster <- function(x, table){
a <- rle(table)
n <- length(a$lengths)
idx <- match(x, a$values, 0)
sequence(a$lengths[idx], cumsum(c(1,a$lengths[-n]))[idx])
}
jon_spring_join_match = function(x, table) {
t <- data.frame(table, index = 1:length(table))
xt <- data.frame(x, index = 1:length(x))
t |>
left_join(xt, join_by(table == x), relationship = 'many-to-many') |>
arrange(index.y) %>%
filter(!is.na(index.y)) %>%
pull(index.x)
}
jared_mamrot_dt <- function(x, table){
table_dt <- data.table(table, index = 1:length(table))
x_dt <- data.table(x, index = 1:length(x))
return(na.omit(table_dt[x_dt, on = .(table == x)][,index]))
}
table = rep(1:1e5, each=10)
x = sample(1:100, 1000, replace = TRUE)
all.equal(onyambu_faster(x, table), jared_mamrot_dt(x, table))
#> [1] TRUE
all.equal(jon_spring_join_match(x, table), jared_mamrot_dt(x, table))
#> [1] TRUE
res <- microbenchmark(onyambu_faster(x, table),
jon_spring_join_match(x, table),
jared_mamrot_dt(x, table),
times = 10)
res
#> Unit: milliseconds
#> expr min lq mean median
#> onyambu_faster(x, table) 38.196317 45.08884 65.22651 52.40748
#> jon_spring_join_match(x, table) 48.697968 74.54407 105.79551 83.11473
#> jared_mamrot_dt(x, table) 9.441176 11.34315 12.99648 11.76324
#> uq max neval cld
#> 64.88688 129.38505 10 a
#> 131.50681 221.16477 10 b
#> 14.05289 21.84779 10 c
autoplot(res)
Created on 2023-10-26 with reprex v2.0.2
1
-
Now I’m curious if duckdb, arrow, or collapse might be even faster…
– Jon Spring11 hours ago
Should be faster as a join. This is >100x faster.
library(dplyr)
join_match = function(x, table) {
t <- data.frame(table, index = 1:length(table))
xt <- data.frame(x, index = 1:length(x))
t |>
left_join(xt, join_by(table == x), relationship = 'many-to-many') |>
arrange(index.y) %>%
filter(!is.na(index.y)) %>%
pull(index.x)
}
Same output, 100-200x as fast, and ~3x as fast as the base R suggestion from @Onyambu (note: that approach has since updated to be similar speed, and a data.table solution is even faster. Using duckdb, or arrow, or collapse to do the join might be faster still. But my observation remains that you can get dramatic speed improvements + legibility by thinking of this as a join):
set.seed(42)
table = rep(1:1e5, each=10)
x = sample(1:100, 1000, replace = TRUE)
bench::mark(
mymatch(x, table) |> as.vector(),
join_match(x, table),
unlist(split(seq(table), table)[as.character(x)],use.names = FALSE),
check = TRUE
)
# A tibble: 3 × 13
expression min median `itr/sec` mem_alloc `gc/sec` n_itr n_gc total_time result
<bch:expr> <bch:tm> <bch:tm> <dbl> <bch:byt> <dbl> <int> <dbl> <bch:tm> <list>
1 as.vector(mymatch(x, table)) 13.8s 13.8s 0.0727 14.9GB 2.83 1 39 13.8s <int>
2 join_match(x, table) 48.7ms 62.2ms 13.8 88.8MB 3.95 7 2 506.3ms <int>
3 unlist(split(seq(table), table)[as.character(x)], use.names = FALSE) 183.6ms 184.5ms 5.31 29.8MB 0 3 0 564.9ms <int>
1
-
mymatchwas producing a matrix. If ok to flatten that to a vector, the output matches my function.– Jon Spring23 hours ago
Using the data in the question this is twice as fast based on the median time on my machine.
table = rep(1:5, each=3)
x = c(2, 5, 2, 6)
mymatch = function(x, table) {
matches = sapply(x, (xx) which(table %in% xx))
unlist(matches)
}
outer_match <- function(x, table) {
z1 <- outer(table, x, "==")
z2 <- z1 * row(z1)
z2[z2 != 0]
}
outer_match(x, table)
## [1] 4 5 6 13 14 15 4 5 6
library(microbenchmark)
microbenchmark(
mymatch(x, table),
outer_match(x, table)
)
## Unit: microseconds
## expr min lq mean median uq max neval cld
## mymatch(x, table) 77.0 79.15 166.696 82.75 84.3 8384.9 100 a
## outer_match(x, table) 35.1 36.75 115.783 41.95 43.1 7410.1 100 a
You can simply run outer + row (short code but might be not that efficient, due to outer), e.g.,
> row(d <- outer(table, x, `==`))[d]
[1] 4 5 6 13 14 15 4 5 6
If the values being matched are integers, then you can use them as index values in a list containing the index values you want (as long as the max integer isn’t so big that the list exceeds your RAM capacity).
# Process the table vector every time
anjama_list <- function(x, table) {
l = vector("list", max(table))
i = 0
for (val in table) {
i = i + 1
l[[val]] = c(l[[val]], i)
}
return(unlist(l[x]))
}
Now, this isn’t as fast as other solutions mentioned here, but since you are reusing the table for multiple lookups, we can precalculate the list creation and reuse it across iterations:
# If the table vector is being reused, only need to process it once
l = vector("list", max(table))
i = 0
for (val in table) {
i = i + 1
l[[val]] = c(l[[val]], i)
}
anjama_list_cache <- function(x, l) {
return(unlist(l[x]))
}
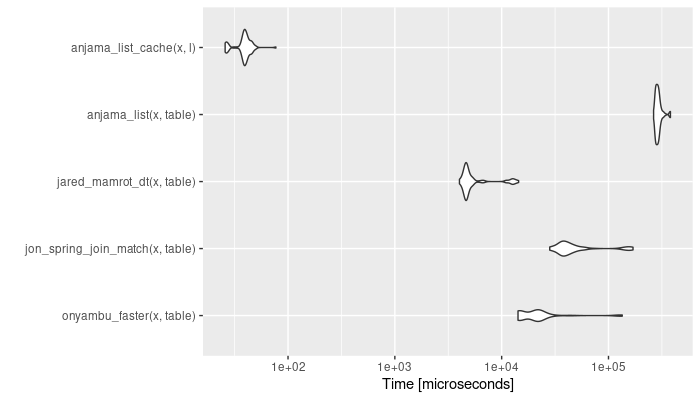
It turns out the list lookup and unlist part is really cheap:
Figure created using the code from jared_mamrot’s answer.
So, it depends on how many times reusing the same table vector makes up for the initial setup. In terms of memory usage, I think the lookup and unlist should also be pretty efficient (the lookup being basically nothing and the unlist being related to the size of your x vector), but I haven’t tried profiling them.








I asked a similar questions some time back: Most efficient way to determine if element exists in a vector
yesterday
When I run
mymatchon the larger sample data, I get a 10×1000 matrix, is that the desired output structure?23 hours ago
@JonSpring – no it was not intended. Well spotted. The desired output format is a simple vector.
23 hours ago
Is table always asorted array of 1:n? with m repetitions or is it a random vector
23 hours ago
@Onyambu No, table is not necessarily 1:n. In my current use case, it's ok to assume it is sorted, although more general solutions are also of interest.
23 hours ago